Recap of OLS
\((Y_i, \pmb{x}_i)\) をrandom vectorとして,\(Y_i \in \mathbb R, \pmb{x}_i \in \mathbb R^k\) とします. 線形回帰では,\((Y_i, \pmb{x}_i)\) について次のようにパラメーターに関して線形な関係があるとして回帰問題を考えます.
\[
\begin{align*}
Y_i &= \pmb{x}_i^T\pmb{\beta} + \epsilon_i \quad (\epsilon_i \text{: errors})
\end{align*}
\]
これを行列表現にした場合,\(\pmb{Y} = (Y_1, \cdots, Y_n)^T\) , \(\pmb{X} = (\pmb{x}_1, \cdots, \pmb{x}_n)^T\) として
\[
\pmb{Y} = \pmb{X}\pmb{\beta} + \pmb{\epsilon}
\]
Assumption: OLSの仮定
mutually independent and identical distribution
\[
(Y_i, \pmb{x_i^T}), i = 1, \cdots, n \text{ は i.i.d}
\]
Linear model
\[
\pmb{Y} = \pmb{X}\pmb{\beta} + \pmb{\epsilon}
\]
error term is conditional-mean zero
\[
\mathbb E[\epsilon_i\vert \pmb{x}_i] = 0
\]
finite moments
\[
\begin{align*}
\mathbb E[Y_i^2] &< \infty\\
\mathbb E[\|\pmb{x}_i^2\|] &< \infty
\end{align*}
\]
Full rank condition
\(\operatorname{rank}(\pmb{X}) = k\) ,つまり完全な多重共線性がない
議論の単純化のため,以下では
\[
\epsilon_1, \cdots, \epsilon_m \overset{\mathrm{iid}}{\sim} N(0, \sigma^2)
\tag{27.1}\]
つまり, Homoscedasticityの仮定の下で話を進めます.
▶ \(\pmb\beta\) の推定
仮定 (2), (3) より \(\mathbb E[Y_i\vert \pmb{x}_i] = \pmb{x}_i^T\pmb{\beta}\) であるので,
\[
\pmb{\beta} = \arg\min_{b} \mathbb E[(Y_i - \pmb{x}_i^T\pmb{b})^2]
\tag{27.2}\]
the joint distribution \((Y_i, \pmb{x}_i)\) は事前には知られていないので、sample analogを用いて以下のように推定します
\[
\begin{align*}
\widehat{\pmb{\beta}}
&= \arg\min_{b} \frac{1}{N}\sum_{i=1}^N(Y_i - \pmb{x}_i^T\pmb{b})^2\\
&= \arg\min_{b} (\pmb{Y} - \pmb{X}\pmb{\beta})^T(\pmb{Y} - \pmb{X}\pmb{\beta})
\end{align*}
\]
上記について,\(b\) のFOCをとると
\[
\pmb{X}^T(\pmb{Y} - \pmb{X}\widehat{\pmb{\beta}}) = 0
\]
仮定 (5) より \(\operatorname{rank}(\pmb{X}) = k\) , つまりfull rankであるので \((\pmb{X}^T\pmb{X})^{-1}\) がとれます.従って,
\[
\widehat{\pmb{\beta}} = (\pmb{X}^T\pmb{X})^{-1}(\pmb{X}^T\pmb{Y})
\tag{27.3}\]
▶ \(\widehat{\pmb{\beta}}\) の漸近分散
population \(\pmb{\beta}\) を用いてerror termを以下のように表します
\[
\begin{align*}
\pmb{Y}_i
&= \pmb{x}_i^T\pmb{\beta} + (\pmb{Y_i} - \pmb{x}_i^T\pmb{\beta})\\
&= \pmb{x}_i^T\pmb{\beta} + \epsilon_i
\end{align*}
\]
両辺に \([\pmb{X}^T\pmb{X}]^{-1}\pmb{X}^T\) をかけると Equation 27.3 より
\[
\widehat{\pmb{\beta}} = \pmb{\beta} + \left[\sum\pmb{x}_i\pmb{x}_i^T\right]^{-1}\sum \pmb{x}_i\epsilon_i
\]
つぎに \(\sqrt{n}(\widehat{{\pmb{\beta}}} - \pmb{\beta})\) で漸近分散を考えると,
\[
\sqrt{n}(\widehat{{\pmb{\beta}}} - \pmb{\beta}) = n\left[\sum\pmb{x}_i\pmb{x}_i^T\right]^{-1}\frac{1}{\sqrt{n}}\sum \pmb{x}_i\epsilon_i
\]
\(\mathbb E[\pmb{x}_ie_i] = 0\) であるので,\(\frac{1}{\sqrt{n}}\sum \pmb{x}_i\epsilon_i\) は location は 0であることがわかります.従って,
\[
\frac{1}{\sqrt{n}}\sum \pmb{x}_i\epsilon_i = \sqrt{n}\left\{\frac{1}{n}\sum \pmb{x}_i\epsilon_i - 0\right\}\overset{\mathrm{d}}{\to} N(0, \mathbb E[\pmb{x}_i\pmb{x}^T\epsilon_i^2])
\]
また,\(\frac{1}{n}\sum\pmb{x}_i\pmb{x}_i^T\) が \(\mathbb E[\pmb{x}_i\pmb{x}_i^T]\) に確率収束するので,スラツキーの定理とCLTにより
\[
\begin{align*}
\sqrt{n}(\widehat{{\pmb{\beta}}} - \pmb{\beta}) \overset{\mathrm{d}}{\to} N(0, \mathbb E[\pmb{x}_i\pmb{x}]^{-1}\mathbb E[\pmb{x}_i\pmb{x}^T\epsilon_i^2]\mathbb E[\pmb{x}_i\pmb{x}]^{-1})
\end{align*}
\]
Equation 27.1 より
\[
\mathbb E[\pmb{x}_i\pmb{x}^T\epsilon_i^2] = \sigma^2\mathbb E[\pmb{x}_i\pmb{x}^T]
\]
が成立するので
\[
\sqrt{n}(\widehat{{\pmb{\beta}}} - \pmb{\beta}) \overset{\mathrm{d}}{\to} N(0, \sigma^2\mathbb E[\pmb{x}_i\pmb{x}]^{-1})
\]
\(\pmb{X}\) で条件付けた分散は
\[
\begin{align*}
\operatorname{Var}(\widehat{\pmb{\beta}}\vert \pmb{X})
& = \mathbb E[(\pmb{X}^T\pmb{X})^{-1}\pmb{X}^T\pmb\epsilon\pmb\epsilon^T\pmb{X}(\pmb{X}^T\pmb{X})^{-1}\vert \pmb{X}]\\
&= (\pmb{X}^T\pmb{X})^{-1}\pmb{X}^T\mathbb E[\pmb\epsilon\pmb\epsilon^T\vert \pmb{X}](\pmb{X}^T\pmb{X})^{-1}\pmb{X}^T\\
&= \sigma^2(\pmb{X}^T\pmb{X})^{-1}
\end{align*}
\]
▶ \(\pmb{Y}\) の条件付き同時分布
Equation 27.1 より\(\pmb{Y}\) の条件付き同時確率密度は
\[
\begin{align*}
p&(Y_1, \cdots, Y_n\vert \pmb{x_1}, \cdots, \pmb{x_n}, \pmb{\beta}, \sigma^2)\\
&= \prod p(Y_i\vert \pmb{x_i}, \pmb{\beta}, \sigma^2) \quad \because\text{i.i.d}\\
&= (2\pi\sigma^2)^{-n/2}\exp\{- \frac{1}{2\sigma^2}\sum_{i=1}^n (Y_i - \pmb{x}_i^T\pmb{\beta})^2\}
\end{align*}
\tag{27.4}\]
つまり,
\[
\{\pmb{Y} \vert \pmb{X}, \pmb{\beta}, \sigma\} \sim \operatorname{N}(\pmb{X}\pmb{\beta}, \sigma^2\pmb{I})
\]
Equation 27.4 の尤度最大化は
\[
\min\sum_{i=1}^n (Y_i - \pmb{x}_i^T\pmb{\beta})^2
\]
を解くことで得られますが,これは Equation 27.2 の問題と対応していることがわかります.
Bayesian Linear Regression
▶ \(\pmb\beta\) の事後分布の導出
\(\pmb{\beta}\) についての事前分布を
\[
\pmb{\beta} \sim \operatorname{N}(\pmb{\beta}_0, \pmb{\Sigma}_0)
\]
事後分布は
\[
\begin{align*}
p&(\pmb\beta\vert \pmb Y, \pmb X, \sigma^2)\\
&\propto p(\pmb Y\vert \pmb X, \pmb \beta, \sigma^2)\times p(\pmb\beta)\\
&\propto \exp\{-\frac{1}{2}(-2\pmb\beta^T\pmb X^T\pmb y/\sigma^2 + \pmb\beta^T\pmb X^T\pmb X\pmb\beta/\sigma^2) - \frac{1}{2}(-2 \pmb\beta^T\pmb\Sigma_0^{-1}\pmb\beta_0 + \pmb\beta^T\pmb\Sigma_0^{-1}\pmb\beta)\} \\
&= \exp\{\pmb\beta^T(\pmb\Sigma_0^{-1}\pmb\beta_0 + \pmb X^T\pmb Y/\sigma^2) - \frac{1}{2}\pmb\beta^T(\pmb\Sigma_0^{-1} + \pmb X^T\pmb X/\sigma^2)\pmb\beta\}
\end{align*}
\]
多変量正規分布を仮定しているので,以下のようにまとめることができます
\[
\begin{align*}
\operatorname{Var}(\pmb\beta\vert\pmb Y, \pmb X, \sigma^2)
&= (\pmb\Sigma_0^{-1} + \pmb X^T\pmb X/\sigma^2)^{-1}\\
\mathbb E[\pmb\beta\vert\pmb Y, \pmb X, \sigma^2]
&= (\pmb\Sigma_0^{-1} + \pmb X^T\pmb X/\sigma^2)^{-1}(\pmb\Sigma_0^{-1}\pmb\beta_0 + \pmb X^T\pmb Y/\sigma^2)
\end{align*}
\tag{27.5}\]
Equation 27.5 について
\[
\begin{gather}
\pmb\beta_0 = \pmb 0\\
\pmb\Sigma_0 = \frac{\sigma^2}{\lambda}\pmb I
\end{gather}
\]
と事前分布のパラメータを設定すると,
\[
\begin{align*}
\mathbb E[\pmb\beta\vert\pmb Y, \pmb X, \sigma^2] = (\lambda\pmb I + \pmb X^T\pmb X)^{-1}\pmb X^T\pmb Y
\end{align*}
\]
となり,次にようにペナルティ \(\lambda\) のRidge repression推定量と一致することがわかります.
Ridge推定量は
\[
\pmb\beta_{ridge} =\arg\min (\pmb Y- \pmb X\pmb \beta)^T(\pmb Y-\pmb X\pmb \beta) + \lambda \|\pmb \beta\|^2
\]
の解と一致します.これについて,FOCをとると
\[
\pmb\beta_{ridge} = (\pmb X^T\pmb X + \lambda \pmb I)^{-1}\pmb X^T\pmb Y
\]
参考までですが,Ridge推定量の条件付き漸近分散は
\[
\begin{align*}
\operatorname{Var}(\pmb\beta_{ridge}\vert \pmb X) &= \sigma^2(\pmb X^T\pmb X + \lambda I_p)^{-1}\pmb X^T\pmb X(\pmb X^T\pmb X + \lambda \pmb I)^{-1}\\
&\leq \sigma^2(\pmb X^T\pmb X)^{-1} = V(\pmb\beta_{ols}\vert \pmb X)
\end{align*}
\]
▶ \(\sigma^2\) の事前分布
多くの正規モデルでは \(\sigma^2\) の準共役事前分布は逆ガンマ分布です. \(\gamma = 1/\sigma^2\) を観測の精度とみなし,
\[
\gamma \sim \operatorname{Ga}(v_0/2, v_0\sigma^2_0/2)
\]
と事前分布を設定すると
\[
\begin{align*}
p&(\gamma\vert\pmb Y, \pmb X, \pmb\beta)\\[5pt]
&\propto p(\gamma)\times p(\pmb Y \vert \gamma, \pmb X, \pmb\beta)\\
&\propto \left[\gamma^{v_0/2 -1} \exp(-\gamma\times v_0\sigma^2_0/2)\right] \times \left[\gamma^{n/2}\exp(-\gamma \times \operatorname{SSR}(\pmb\beta)/2)\right]\\
&= \gamma^{(v_0+n)/2 -1} \exp(-\gamma [v_0\sigma^2_0 + \operatorname{SSR}(\pmb\beta)]/2)\\[5pt]
&\text{where }\quad \operatorname{SSR}(\pmb\beta) = (\pmb Y - \pmb X\pmb\beta)^T(\pmb Y - \pmb X\pmb\beta)
\end{align*}
\]
これはガンマ分布とみなせるので,\(\gamma = 1/\sigma^2\) より,事後分布は
\[
\{\sigma^2\vert\pmb Y, \pmb X, \pmb\beta\} \sim \operatorname{inverse-gamma}((v_0+n)/2, [v_0\sigma^2_0 + \operatorname{SSR}(\pmb\beta)]/2)
\]
従って,ベイジアンアップデートは以下のような手順で行います.
▶ Step 1: \(\pmb\beta\) の更新
\(\pmb V = \operatorname{Var}(\pmb\beta\vert \pmb Y, \pmb X, \sigma^{2}_{(s)}), \pmb m = \mathbb E[\pmb\beta\vert \pmb Y, \pmb X, \sigma^{2}_{(s)}]\) の計算\(\pmb\beta_{(s+1)}\) を次のように生成
\[
\pmb\beta_{(s+1)}\sim \operatorname{N}(\pmb m, \pmb V)
\]
▶ Step 2: \(\sigma^2\) の更新
\(\operatorname{SSR}(\pmb\beta_{(s+1)})\) を計算\(\sigma^{2}_{(s+1)}\) を次のように生成
\[
\sigma^{2}_{(s+1)}\sim \operatorname{inverse-gamma}((v_0+n)/2, [v_0\sigma^2_0 + \operatorname{SSR}(\pmb\beta_{(s+1)})]/2)
\]
Bayesian Linear Regression with JAX
Code
import numpy as npimport pandas as pdimport matplotlib.pyplot as pltimport statsmodels.api as smfrom sklearn import linear_modelfrom regmonkey_style import stylewizard as swfrom jax import numpy as jnp, scipy as jscipyfrom jax import gradimport scipy.stats
Code
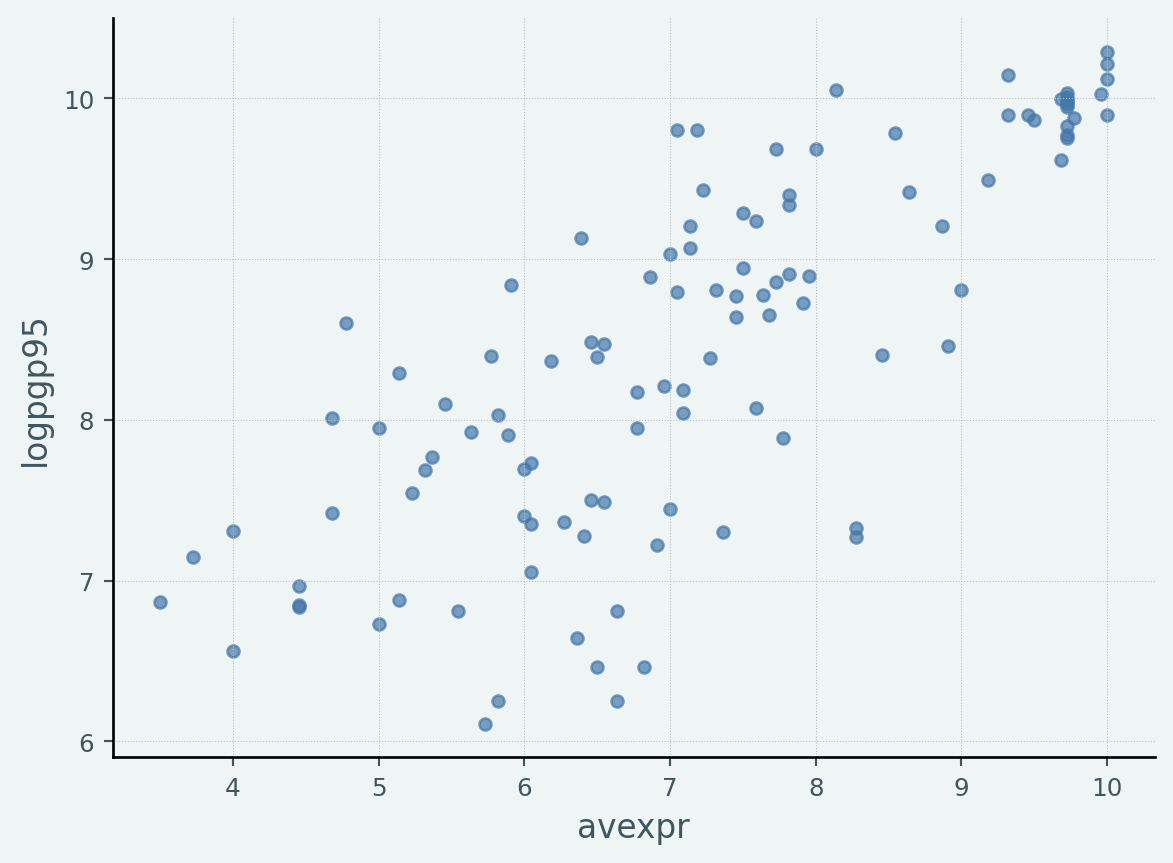
= pd.read_stata("https://github.com/QuantEcon/lecture-python/blob/master/source/_static/lecture_specific/ols/maketable1.dta?raw=true" = ["logpgp95" , "avexpr" ])
1
AGO
8.000000
1.0
5.363636
7.770645
3.0
3.0
0.0
1.0
280.000000
5.634789
-3.411248
1.0
2
ARE
0.000000
1.0
7.181818
9.804219
NaN
NaN
NaN
NaN
NaN
NaN
NaN
NaN
3
ARG
60.000004
1.0
6.386364
9.133459
1.0
6.0
3.0
3.0
68.900002
4.232656
-0.872274
1.0
5
AUS
98.000000
1.0
9.318182
9.897972
7.0
7.0
10.0
7.0
8.550000
2.145931
-0.170788
1.0
6
AUT
100.000000
0.0
9.727273
9.974877
NaN
NaN
NaN
NaN
NaN
NaN
-0.343900
NaN
Code
"regmonkey_scatter" )= 'avexpr' , y= 'logpgp95' , kind= 'scatter' )
▶ MLE
Code
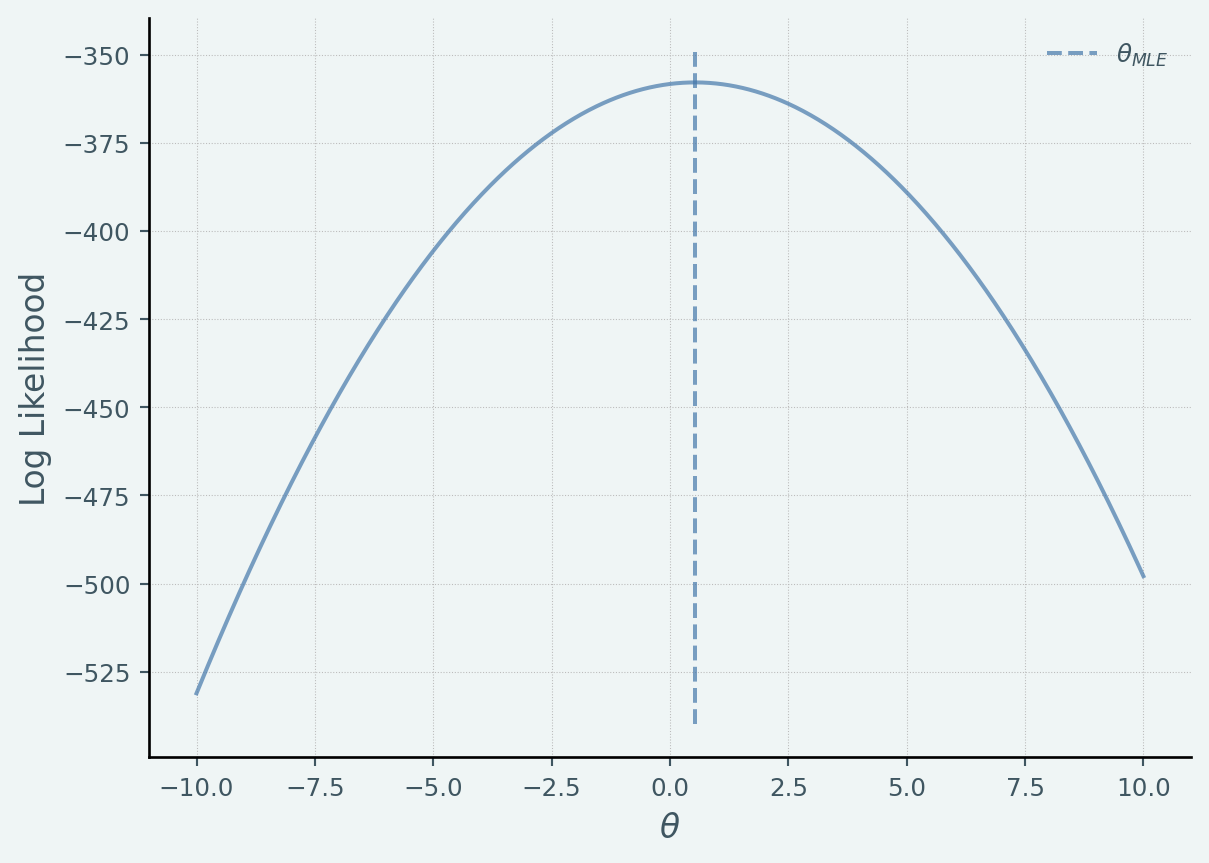
def LogLikelihood(y, x, theta):= x.shape[0 ]return np.log(scipy.stats.multivariate_normal.pdf(y.ravel(), (theta* x).ravel(), np.eye(N)* sigma_n** 2 ))= df['avexpr' ].values= df['logpgp95' ].values# FWL = x - np.mean(x)= y - np.mean(y)# MLE 42 )= 10 = np.linspace(- 10 ,10 ,1000 )= [LogLikelihood(y_mle, x_mle, t1) for t1 in T]= plt.subplots()= np.sum (x_mle* y_mle)/ np.sum (np.square(x_mle))* ax.get_ylim(), linestyle= '--' , label= '$ \\ theta_ {MLE} $' )"$ \\ theta$" ); ax.set_ylabel("Log Likelihood" ); = [1 ,1 ]); print ("MLE beta: {} " .format (beta_mle))
MLE beta: 0.5318712592124939
▶ OLS
Code
= df.copy()'const' ] = 1 = sm.OLS(endog= df_ols['logpgp95' ], exog= df_ols[['const' , 'avexpr' ]], \ = 'drop' )= reg_ols.fit()print (results.summary())
OLS Regression Results
==============================================================================
Dep. Variable: logpgp95 R-squared: 0.611
Model: OLS Adj. R-squared: 0.608
Method: Least Squares F-statistic: 171.4
Date: Sat, 19 Oct 2024 Prob (F-statistic): 4.16e-24
Time: 10:17:22 Log-Likelihood: -119.71
No. Observations: 111 AIC: 243.4
Df Residuals: 109 BIC: 248.8
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 4.6261 0.301 15.391 0.000 4.030 5.222
avexpr 0.5319 0.041 13.093 0.000 0.451 0.612
==============================================================================
Omnibus: 9.251 Durbin-Watson: 1.689
Prob(Omnibus): 0.010 Jarque-Bera (JB): 9.170
Skew: -0.680 Prob(JB): 0.0102
Kurtosis: 3.362 Cond. No. 33.2
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
▶ Rdige
Code
= sm.OLS(endog= df_ols['logpgp95' ], exog= df_ols[['const' , 'avexpr' ]], \ = 'drop' )= reg_l2.fit_regularized(L1_wt= 0. , alpha= [1 / 2 ,1 / 2 ])
array([0.55745346, 1.05775113])
Code
= sm.OLS(endog= y_mle, exog= x_mle, \ = 'drop' )= reg_l2.fit_regularized(L1_wt= 0. , alpha= [1 ])
Code
from sklearn.linear_model import Ridgefrom sklearn.linear_model import ridge_regression= ridge_regression(x.reshape(- 1 ,1 ), y, alpha= 1.0 , return_intercept= True )print (coef, intercept)
▶ bayesian regression
Code
from IPython.display import set_matplotlib_formatsimport seaborn as snsfrom jax import random, vmapimport jax.numpy as jnpfrom jax.scipy.special import logsumexpimport numpyrofrom numpyro import handlersfrom numpyro.diagnostics import hpdiimport numpyro.distributions as distfrom numpyro.infer import MCMC, NUTSdef model(x= None , y= None ):#a = numpyro.sample("a", dist.Normal(0.0, 1)) = 0.0 , 0.0 if x is not None := numpyro.sample("bM" , dist.Normal(0.0 , 1 ))= bM * x= numpyro.sample("sigma" , dist.Gamma(1.0 ))#mu = a + M = M"obs" , dist.Normal(mu, sigma), obs= y)# Start from this source of randomness. We will split keys for subsequent operations. = random.PRNGKey(0 )= random.split(rng_key)= NUTS(model)= 200 = MCMC(kernel, num_warmup= 100 , num_samples= num_samples)= x_mle, y= y_mle= mcmc.get_samples()
mean std median 5.0% 95.0% n_eff r_hat
bM 0.53 0.04 0.53 0.46 0.58 166.41 1.00
sigma 0.72 0.05 0.72 0.65 0.81 50.98 1.00
Number of divergences: 0
def model(x= None , y= None ):= numpyro.sample("a" , dist.Normal(5.0 , 1 ))= 0.0 , 0.0 if x is not None := numpyro.sample("bM" , dist.Normal(0.0 , 1 ))= bM * x= numpyro.sample("sigma" , dist.InverseGamma(1.0 ))= a + M"obs" , dist.Normal(mu, sigma), obs= y)# Start from this source of randomness. We will split keys for subsequent operations. = random.PRNGKey(42 )= random.split(rng_key)= NUTS(model)= 2000 = MCMC(kernel, num_warmup= 1000 , num_samples= num_samples)= x, y= y= mcmc.get_samples()
mean std median 5.0% 95.0% n_eff r_hat
a 4.67 0.29 4.66 4.22 5.15 647.40 1.00
bM 0.53 0.04 0.53 0.46 0.59 649.07 1.00
sigma 0.72 0.05 0.72 0.65 0.80 935.07 1.00
Number of divergences: 0
Code
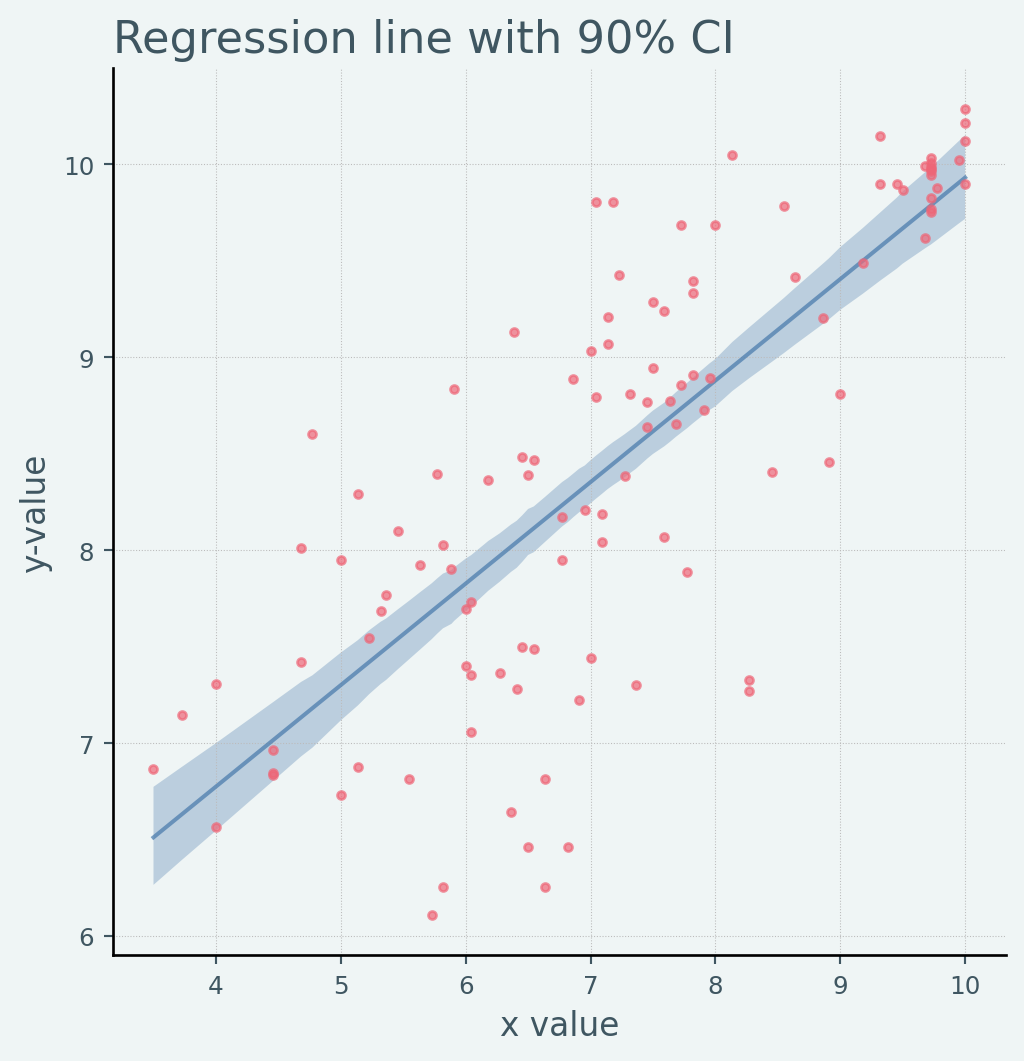
def plot_regression(x_data, y_mean, y_hpdi, y_data):# Sort values for plotting by x axis = jnp.argsort(x_data)= x_data[idx]= y_mean[idx]= y_hpdi[:, idx]= y_data[idx]# Plot = plt.subplots(nrows= 1 , ncols= 1 , figsize= (6 , 6 ))"o" )0 ], hpdi[1 ], alpha= 0.3 , interpolate= True )return ax# Compute empirical posterior distribution over mu = ("a" ], - 1 )+ jnp.expand_dims(samples_1["bM" ], - 1 ) * x= jnp.mean(posterior_mu, axis= 0 )= hpdi(posterior_mu, 0.9 )= plot_regression(x, mean_mu, hpdi_mu, y)set (= "x value" , ylabel= "y-value" , title= "Regression line with 90% CI" ;
Code
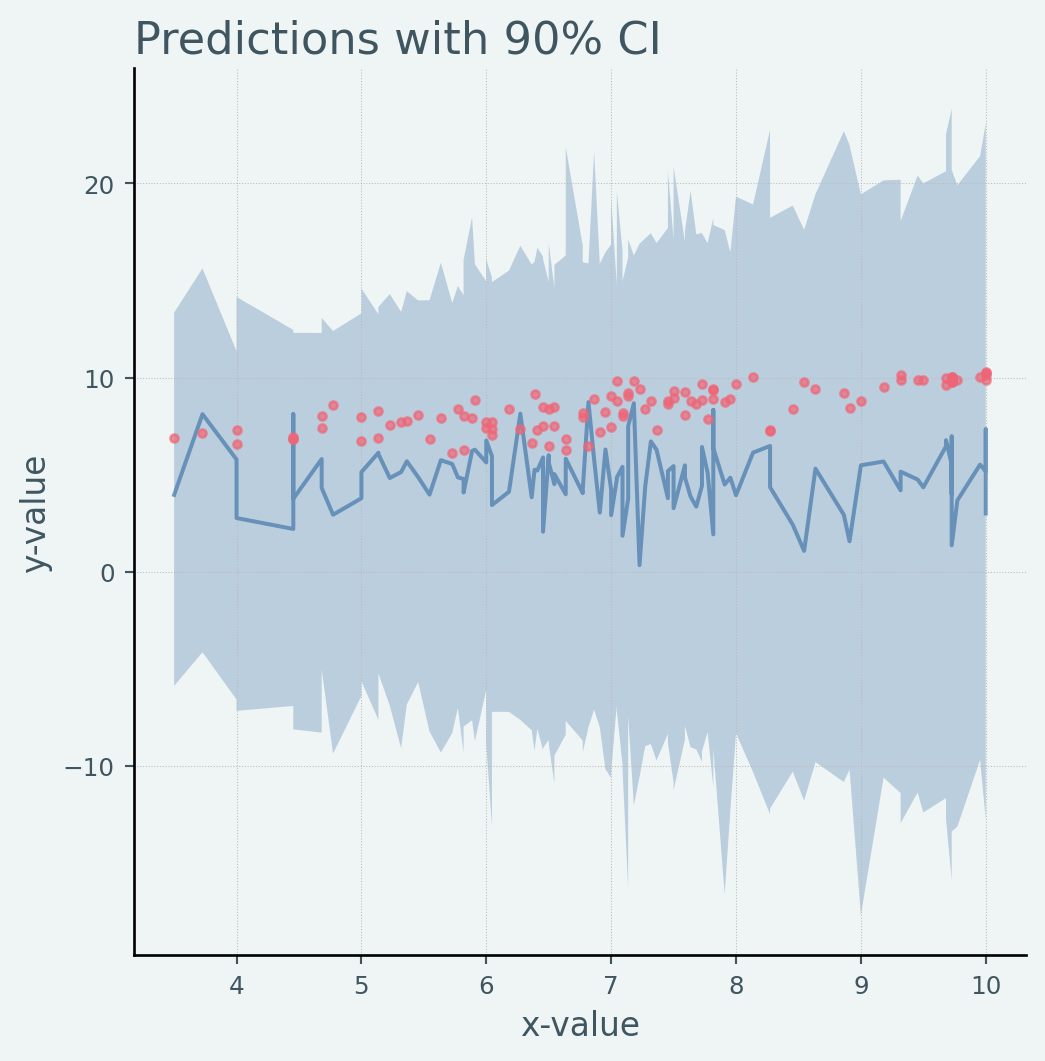
from numpyro.infer import Predictive= random.split(rng_key)= Predictive(model, num_samples= 100 )= prior_predictive(rng_key_, x)["obs" = jnp.mean(prior_predictions, axis= 0 )= hpdi(prior_predictions, 0.9 )= plot_regression(x, mean_prior_pred, hpdi_prior_pred, y)set (xlabel= "x-value" , ylabel= "y-value" , title= "Predictions with 90% CI" );
Code
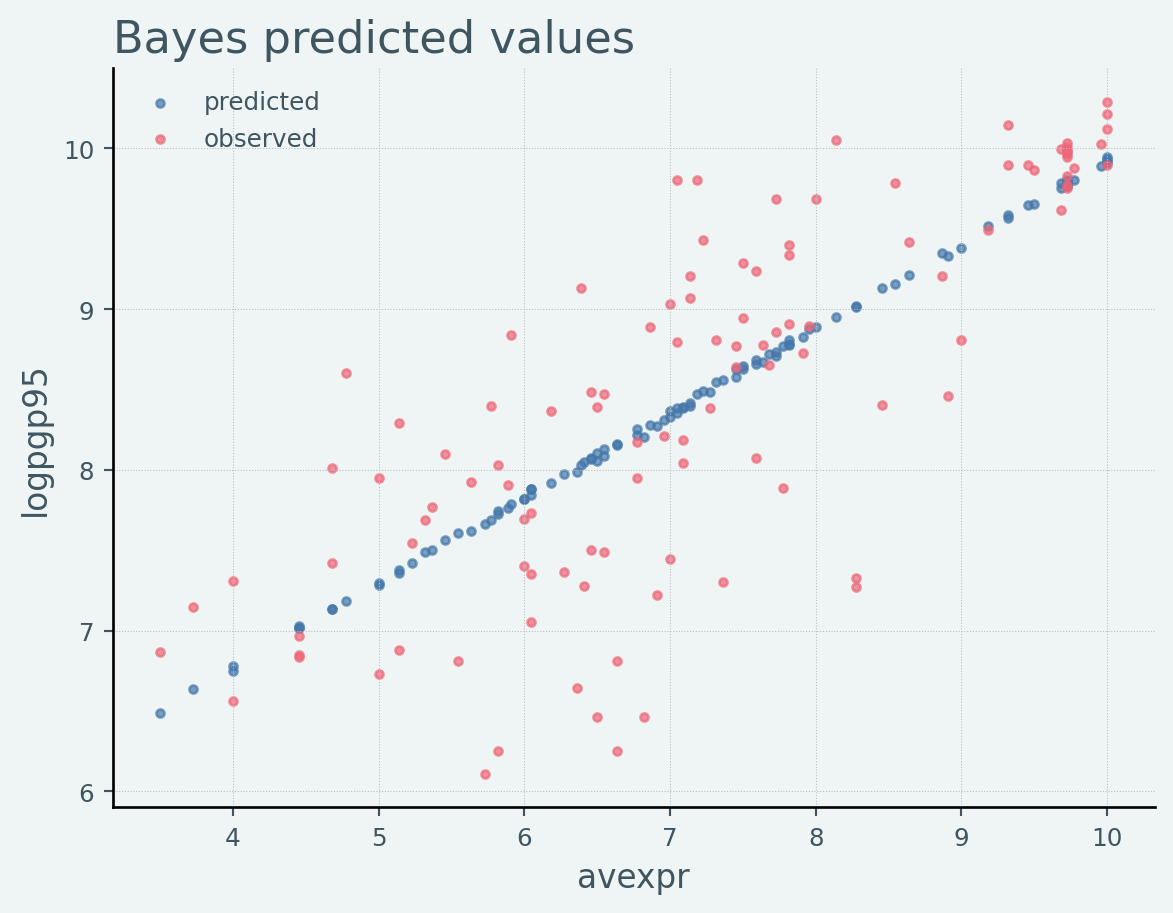
= df.copy()= random.split(rng_key)= Predictive(model, samples_1)= predictive(rng_key_, x= x)["obs" ]"Mean Predictions" ] = jnp.mean(predictions, axis= 0 )"shortnam" , "logpgp95" , "Mean Predictions" ]].head()
1
AGO
7.770645
7.498785
2
ARE
9.804219
8.472743
3
ARG
9.133459
8.029100
5
AUS
9.897972
9.564693
6
AUT
9.974877
9.770352
Code
# Plot predicted values = plt.subplots()'avexpr' ], df_bayes["Mean Predictions" ],= 'predicted' )# Plot observed values 'avexpr' ], df_bayes["logpgp95" ],= 'observed' )= 'upper left' )'Bayes predicted values' )'avexpr' )'logpgp95' )
Code
def log_likelihood(rng_key, params, model, * args, ** kwargs):= handlers.condition(model, params)= handlers.trace(model).get_trace(* args, ** kwargs)= model_trace["obs" ]return obs_node["fn" ].log_prob(obs_node["value" ])def log_pred_density(rng_key, params, model, * args, ** kwargs):= list (params.values())[0 ].shape[0 ]= vmap(lambda rng_key, params: log_likelihood(rng_key, params, model, * args, ** kwargs)= log_lk_fn(random.split(rng_key, n), params)return (logsumexp(log_lk_vals, 0 ) - jnp.log(n)).sum ()= random.split(rng_key)print ("Log posterior predictive density: {} " .format (= x,= y,
Log posterior predictive density: -119.95238494873047